Quickstart
Quickstart
Section titled “Quickstart”FBTK allows you to go from a simple SMILES string to a fully relaxed bulk system in just a few lines of code.
Quick construction from SMILES
Section titled “Quick construction from SMILES”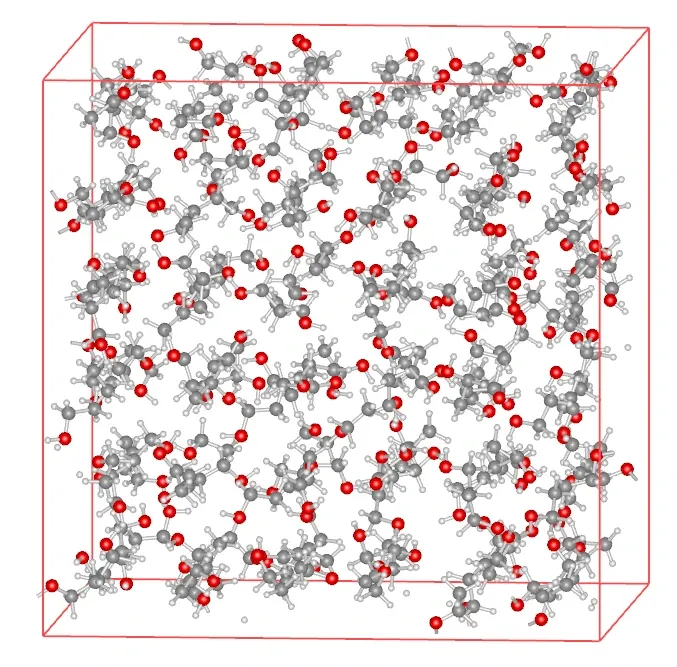
This example creates a bulk system containing Ethanol and Water, relaxes the structure to remove overlaps, and exports it for MD simulation.
import fbtk
# 1. Define Moleculeethanol = fbtk.Molecule.from_smiles("CCO", name="Ethanol")
# 2. Build Systembuilder = fbtk.Builder(density=0.789) # Specify target density (g/cm3)builder.add_molecule(ethanol, count=200)system = builder.build()
# 3. Structural Relaxation (Overlap Removal)system.relax() # 1000 steps (default)
# 4. Save the result (.mol2 preserves unit cell information)system.to_file("initial.mol2")
# (Optional) Export to ASE# atoms = system.to_ase()# atoms.write("initial.data", format="lammps-data")What’s Happening?
Section titled “What’s Happening?”- Smart Packing: Molecules are randomly placed and rotated while avoiding severe overlaps.
- Internal 3D Generation: 3D structures for “Ethanol” and “Water” are generated on-the-fly from SMILES using the internal VSEPR/UFF engine.
- Fast Relaxation: The
relax()method uses a high-performance Rust implementation of the Universal Force Field (UFF) and the FIRE optimizer to stabilize the system in milliseconds. - Format Interoperability: The resulting
Systemobject can be converted to an ASEAtomsobject or exported to a.mol2file (complete with unit cell parameters and connectivity) for use in AmberTools or LAMMPS.
Next Steps
Section titled “Next Steps”- System Building: Learn how to create polymers and more complex mixtures.
- Analysis: Learn how to compute RDF and MSD at high speed.
- CLI: Run builds and analysis directly from the command line without Python code.